Tetra Benchling Pipeline
The Tetra Benchling Pipeline pushes instrument and experimental results to Benchling. This integration component uses Tetra Data Pipelines to push data to Benchling by using DataWeave and the Benchling Python software development kit (SDK).
NOTE
For setups that require bi-directional communication between the Tetra Data Platform (TDP) and Benchling, see the Tetra Benchling Connector v1 deployment option. For more information, see Tetra Benchling Integration.
Design Overview
The Tetra Benchling Pipeline pushes instrument and experimental results to three different object constructs within Benchling:
Custom Entities: The construct to hold information that pertains to a particular user-defined entities like samples, batches, injections, etc. Custom Entities are part of Benchling’s Registry.Runs: The construct to hold information pertaining to an experimental run performed through an instrument, robot, or software analysis pipeline.Results: Structured tables that allow you to capture experimental or assay data. Results can be linked with Custom Entities in the Registry or to a Run in a notebook entry.
For more information, see Benchling’s developer platform documentation.
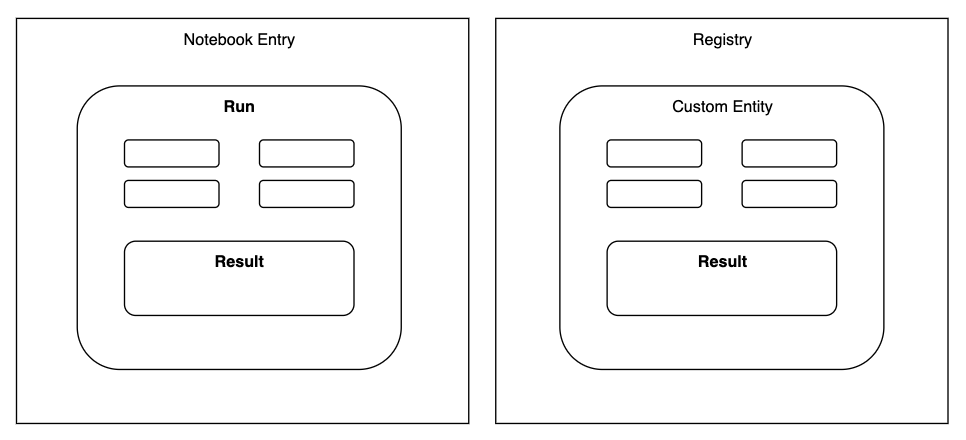
Figure 1. Object constructs and their relationships in Benchling
The Tetra Benchling Pipeline uses Tetra Data Pipelines to push data to Benchling by using DataWeave and the Benchling Python software development kit (SDK). DataWeave scripts as a configuration input allows for scientific data engineers to configure their pipelines to match how they have set up the data schemas in Benchling without worrying about the underlying code or infrastructure of the integration mechanism.
Integration Modalities
Where can data go?
The TetraScience integration to Benchling utilizes the Benchling Python SDK and supports data integration to various Benchling Applications and endpoints. Some examples of such are:
- Creating a Benchling Lab Automation Run and adding experimental information and images to the Run, making it available for a Benchling user to insert the Run from the Benchling Notebook Inbox.
- Creating Benchling Results to record data such as plate reader results or a table recording a chromatography run’s fractions or peaks results, and linking this Result to a Lab Automation Run or a Custom Entity in Benchling Registry, or both.
Benchling File Output Processor for Large Data Set Support
If a large data set from the Tetra Data Lake is desired to be pushed to Benchling, our integration also supports the use of Benchling’s Output File Processor. This is the preferred approach in order to reduce the chances of hitting the API rate limit for uploading Results to Benchling. However, given that the Output Processor will process the given CSV file and generate one Results table, every data push will result in a new Results table. This may not be the desired behavior depending on the scientific use case and how data is being ingested into the Tetra Data Lake and is desired to be processed.
See the Modalities Decision Tree section for examples of how different scientific use cases result in different modalities of sending data to Benchling that the TetraScience integration facilitates.
Integration Modalities Decision Tree
Our integration with Benchling Notebook uses Benchling Lab Automation to facilitate automated data and result transfer, reducing manual transcription.
If Benchling Lab Automation is not used, the TetraScience integration to Benchling only supports pushing Results to Custom Entities in Benchling Registry.
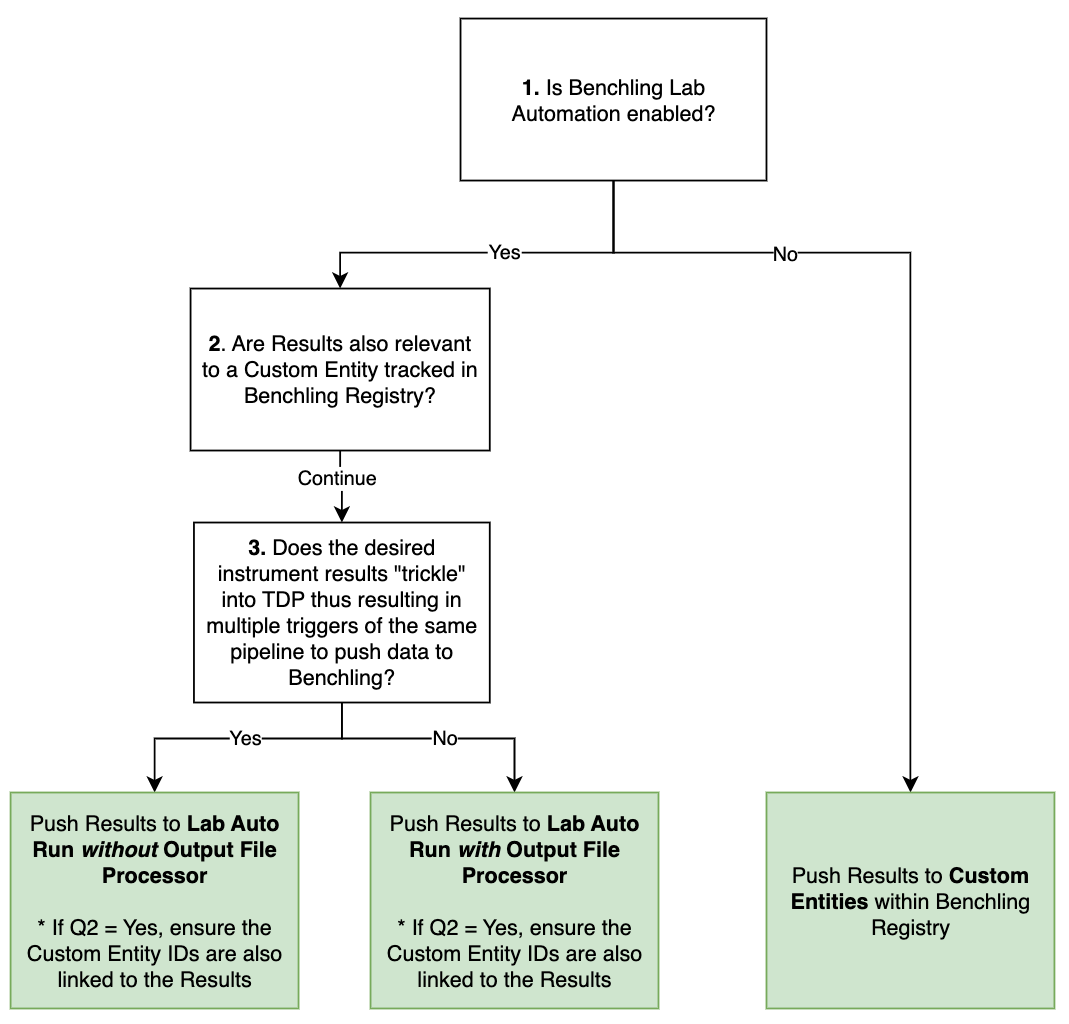
Figure 2. Decision Tree to assist in how to push data to Benchling from the Tetra Data Platform
Below are examples of scientific use cases and how the integration approach follows the modalities decision tree.
Example 1: Bioreactor Run
Scientific use case summary
A bioreactor run typically occurs for 30-90 days and throughout this run, the process can be monitored through the bioreactor control system PC. For run status updates and data sharing, scientists typically want a daily report out of minimum value, maximum value, mean, median, standard deviations, any alarm triggers, and cumulative values (if appropriate) for critical process parameters such as oxygen gas flow rate, stir rate, media temperature.
This type of run information and process data logging into a notebook entry allows scientists to document their bioreactor run for due-diligence, for record, as well as for quick snapshot assessments if the run is going as planned or not.
Recommended integration approach
Process data will likely be entering into the Tetra Data Lake in batches throughout the full bioreactor run. As data is received and processed in batches, Tetra Pipelines can be used to perform data transformation and basic calculations as appropriate to match the defined schemas of the Benchling Notebook Entry Run and Result.
We recommend not using the Output File Processor so that every new result entry can be appended to the same Results table in the Run.
If the particular cell line or cell batch is captured in Benchling Registry, then the generated Results that are in the Run can also be associated with the relevant Registry Custom Entities.
Example 2: Results From a Plate Reader
Scientific use case summary
A microplate reader is used to measure absorbance, fluorescence and luminescence from a 96-well plate to quantify protein expression. Scientists may want to see the result tables structured in various ways: a tabular representation of the plate and one table per measurement type, or perhaps all measurement values shown in one table per sample correlated with the different dilutions of that sample. However, typically each sample would be tracked in Benchling Registry and results would need to be associated back to the Custom Entities.
Recommended integration approach
Plate Reader results can be large in size, thus using the Output File Processor is recommended. Especially for any dataset coming from plates with greater than 24 wells, using the Output File Processor is strongly encouraged to not hit any Benchling API limits.
Additionally, if association of results to Custom Entities is desired, using the Output File Processor can also facilitate creating and registering Custom Entities.
Benchling Authentication
The TetraScience integration to Benchling supports both the Benchling App Authentication as well as API Key Authentication. We recommend saving any authentication credentials in the Tetra Data Platform’s (TDP) Shared Settings and Secrets storage.
App Authentication
When using app authentication, TetraScience will authenticate as an app in Benchling using the provided client ID and client secret. Actions performed by TetraScience will be governed by the permissions of the app that corresponds to the credentials. This is the preferred method of integrating TDP with Benchling, allowing more granular permissions and easy identification of API-driven activity in the logs. More information is available at Getting Started with Benchling Apps .
API Key Authentication
When using API key authentication, TetraScience will authenticate as a normal user in Benchling using the provided API key. Actions performed by TetraScience will be governed by the permissions of the user that corresponds to the API key. This is easy to get started with but not preferred for a production solution.
Setting up SSPs with the Tetra Benchling Pipeline
The TetraScience to Benchling integration is powered by Tetra Data pipelines. The protocol that is used for sending data from TDP to Benchling is common/ids-to-benchling.
For advanced users that are interested in developing self-service Tetra Data pipelines (SSPs), the underlying productized task-scripts common/dataweave-util and common/benchling-util can be leveraged to build your SSP.
Protocol Steps
The most up-to-date information and technical specifications are in the Readme for the protocol common/ids-to-benchling.
Refer to READMEs on TDP for Latest Implementation Docs
Refer to the README for the protocol
common/ids-to-benchlingfor the latest technical specifications and details. This can be accessed on TDP.The below information is written referencing
common/ids-to-benchling:v3.0.0
The ids-to-benchling protocol is comprised of two steps:
-
Transform with DataWeave: This setup allows for the custom transformation of the contents and attributes of the input file that triggered the pipeline. The transformation constructs the exact schema structure and content to be sent to the targeted Benchling objects.Refer to the protocol’s README on TDP for more details about the expected output object structure that is to be fed into the next protocol step.
-
Push to Benchling: This step will take the transformed input from Step 1 and parse it to perform the appropriate Benchling SDK calls for sending the provided contents to the Benchling Assay Runs or Benchling Registry Custom Entities.
Note that the Benchling Project ID (
projectId) is required for pushing data to a Benchling Notebook Entry.
About DataWeave
Mulesoft DataWeave is a functional language for performing transformations between different representations of data. As we use it, it can take XML, CSV or JSON data and convert it to one of these formats. This gives scientific data engineers a powerful tool to programmatically tailor exactly how their data in the TDP should be mapped to specific schemas defined within their Benchling environment.
DataWeave Interface and Reference in Pipeline Configuration
A DataWeave file with extension .dwl can be manually uploaded to TDP. This file can be referenced in the integration pipeline configuration with the following information:
- File’s full file path on TDP:
file.pathfield provided in “File Information” - File’s version on TDP:
file.versionfield provided in ”File Information”
Recommendations for Protocol Configuration
- We recommend setting the
Retry Behaviorfor this integration pipeline to be0. The reason being that if an error occurs, it is likely due to a malformed JSON payload, which can result in unexpected data in Benchling. This can easily be cleaned up on Benchling by archiving results, but failed retries can lead to additional clean up steps for the end user. - We recommend using Benchling App Authentication for protocol configuration whenever possible.
Updated over 1 year ago
